This project is a labor of passion: a fusion of my clinical background as a pharmacist and my growing passion for data analysis.
My goal was simple: to use data to identify which crucial Monoclonal Antibodies (mAbs) are missing from the Thai market despite being available as affordable biosimilars in the US.
I’ve combined storytelling and data visualization to summarize this journey. Each page represent a core step in the analytical process, moving from raw data to actionable clinical insights. I hope you find this process as fascinating as I did!
How I made this?
Inspiration
As a pharmacist in Thailand, I see the innovation lag firsthand. Advanced biologic therapies (mAbs) are transforming medicine, but their high costs often keep them them out of reach for most Thai patients. While, In the US, Biosimilars, a safe, interchangeable , affordable versions of these drug launch as soon as patents expire.
I wanted to find the answer: Which critical molecules are we missing in Thailand that are already available in the US?
Methodology: Data analysis
1. Data preparation
1.1 Prepare US mAbs data from Purple Book
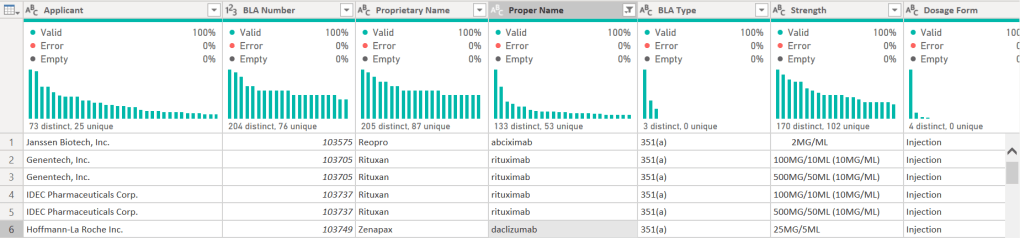
I started with the US FDA Purple Book. I prefer using the data transformation pane in Power BI for initial exploration. The automatic column profiles and quality distributions give me an instant glimpse of the data.
I cleaned the generic name (Proper Names) by stripping manufacturer suffixes (e.g., changing “Adalimumab-atto” to “Adalimumab”) to create a clean list for cross-referencing.
A part of the head of data:
the table schema:
1.2 Prepare Thai mAbs data from Thai FDA scraping
While the US data was a simple CSV, the Thai data was a different story. Since no public dataset existed, I built a Scrapy crawler in Python to navigate the Thai FDA search portal and build a mirrored dataset.
- I used Google Colab and Pandas to manage my search list, then deployed the Scrapy crawler to capture product names, license holders, and registration statuses.
- I used Power BI to join these two datasets. This allowed me to identify “mismatches” which are mAbs registered in the US but absent in Thailand.
My codes for scraping are:
%%writefile med_spider.py
import scrapy
class MedSpider(scrapy.Spider):
name = "meds"
# 1. Added a User-Agent
custom_settings = {
'USER_AGENT': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) Chrome/119.0.0.0 Safari/537.36',
'COOKIES_ENABLED': True, # ASP.NET sites NEED cookies to track your session
'DOWNLOAD_DELAY': 5, # Wait 5 seconds between drugs
'CONCURRENT_REQUESTS': 1, # One at a time to stay under the radar
'USER_AGENT': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) Chrome/119.0.0.0 Safari/537.36',
'FEEDS': {
'med_results.csv': {
'format': 'csv',
'encoding': 'utf-8-sig',
'overwrite': True,
}
}
}
start_urls = ['Thai FDA drug searching URL']
usmed_list = [list of mAb generic names from Purple Book]
# 1. This replaces the default start_urls behavior
def start_requests(self):
for drug in self.usmed_list:
# visit the landing page once for each drug to get a fresh ViewState
yield scrapy.Request(
url=self.start_urls[0],
callback=self.parse,
meta={'search_term': drug}, # save the drug name
dont_filter=True # for hitting the same URL multiple times
)
def parse(self, response):
# retrieve the drug name saved in meta
drug_to_search = response.meta['search_term']
# from_response: handle the hidden ASP.NET ViewState
return scrapy.FormRequest.from_response(
response,
formdata={
'ctl00$ContentPlaceHolder1$txt_substance': drug_to_search,
'ctl00$ContentPlaceHolder1$btn_sea_drug': 'ค้นหา'
},
callback=self.parse_results,
meta={'search_term': drug_to_search} # Pass it forward again to the results
)
def parse_results(self, response, ):
search_term = response.meta['search_term']
rows = response.css('tr.rgRow, tr.rgAltRow')
self.logger.info(f"Found {len(rows)} rows on the page!") # show logs
for row in rows:
# gets text inside <span> or <a> tags.
data = row.css('td:not([style]) ::text').getall()
# Clean up the list (keep empty strings)
clean_data = [item.strip() if item else "" for item in data]
if len(clean_data) > 0:
yield {
'searched_drug': search_term,
'registration_no': clean_data[1] if len(clean_data) > 0 else None,
'trade_name': clean_data[3] if len(clean_data) > 3 else None,
'licensee': clean_data[4] if len(clean_data) > 4 else None,
'drug_type': clean_data[5] if len(clean_data) > 5 else None,
'status': clean_data[7] if len(clean_data) > 5 else None
}
!scrapy runspider med_spider.py
1.3 Identifying hidden gaps
Initial results showed that most of absent drugs were orphan drugs or for rare diseases. To find the “real” opportunities, I needed more information. These are what I started with:
- Market Demand: Top 20 Global Sales data (via Pharmashots) to identify clinical demand and physician trust worldwide.
- Prevalence data: Which disease are crucial in Thailand?
- HDC website shows prevalence data of crucial diseases with ICD-10 code related to those diseases (2025).
- It also show diseases in “Service plan” which are the group of diseases that intensively monitored by Thailand’s Ministry of Public Health (MOPH).
- WHO Top cause of death of Thai population (2021).
- Reimbursement Barriers: National List of Essential Medicines (NLEM) is the “optimum list” of fundamental treatments. It serves as the official reference for reimbursement across public health insurance scheme. there are 7 mAbs in this list.
- Biosimilar gap: I used “BLA type” column, a original/biosimilar label from Purple Book to evaluate a status of Thai mAbs list.
2. Modeling
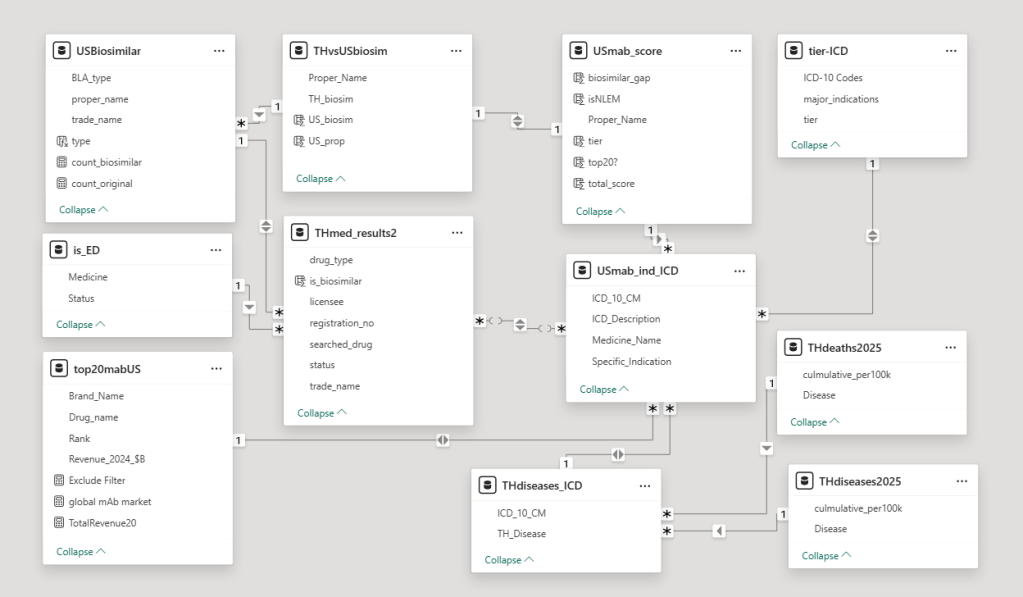
Building this database was an iterative process. This dataset is not perfect. It’s grew organically as I added data one-by-one, but it taught me lessons for my next project:
- Whenever possible, collect all data before designing the schema.
- Establishing strict naming rules early to prevent inconsistent naming.
- Moving toward a star schema and database normalization makes the system much more robust as data piles up.
3. Visualization
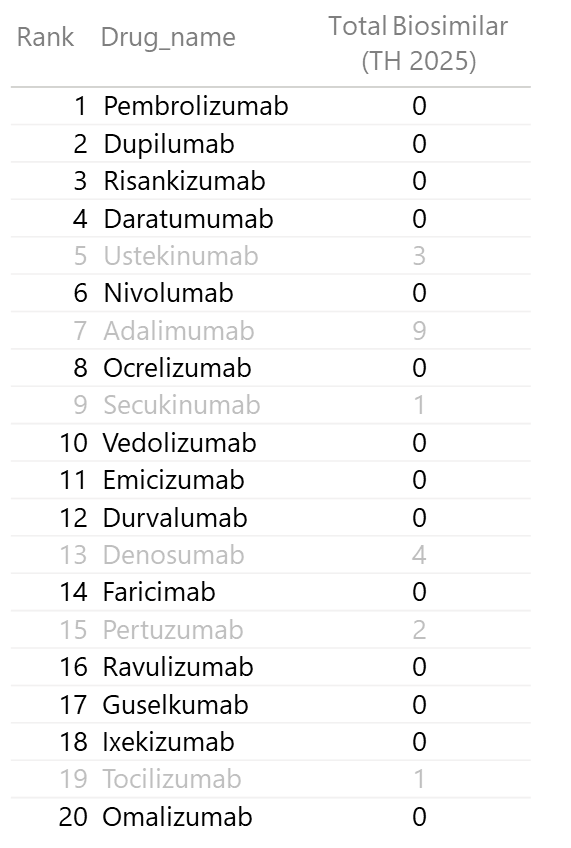
3.1 Top 20 Monoclonal Antibodies by Global Sales
I discovered that the Top 20 mAbs account for 60% of total global mAb sales. This is a massive concentration of value!
By highlighting which of these 20 have zero biosimilar competition in Thailand, I identified the most significant market gaps.

3.2 Clinical Impact
Prevalence data can be “noisy”. Disease are grouped to a big categories like “heart disease” consisting of many ICM-10 codes which make the prevalence illogically high. So, I came up with the decision tree for categorizing each mAbs in to three tiers, inspired by inclusion criteria of the NELM. Categorization was derived from the MOPH 2025 National Health Priorities and the HITAP Burden of Disease framework(Issue 26).
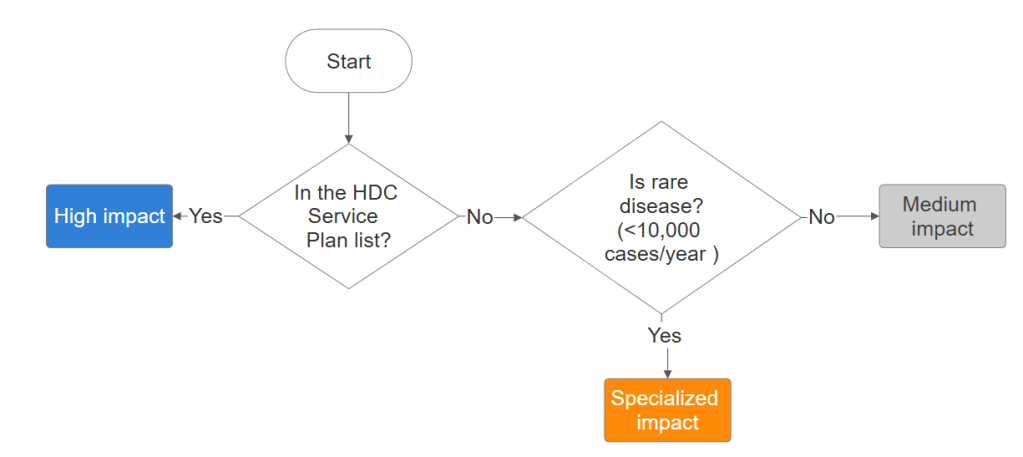
- Tier 3 (High): Aligned with the Thai MOPH “Service Plan” (e.g., Cancer, Stroke).
- Tier 2 (Medium): Prevalence is higher that rare disease threshold (>10,000 case/year).
- Tier 1 (Specialized): The leftovers: Rare or specialized indications.
3.3 NLEM gap
I created bar chart showing count of each mAbs in NLEM list biosimilars in Thailand. The data show 2 mAbs don’t have biosimilar yet, which is a huge market gap.
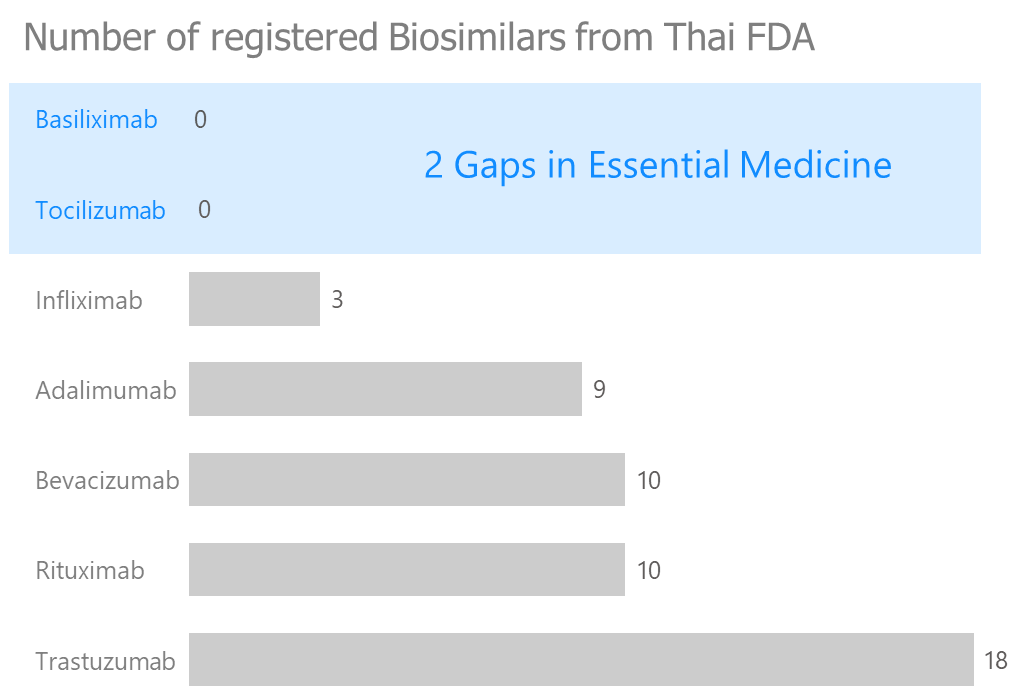
3.4 Biosimilar availability gap
I compared the number of US biosimilars (Blue for positivity/opportunity) against Thai biosimilars (Orange for competition/saturation). This visual logic allowed me to assign scores for my final index.
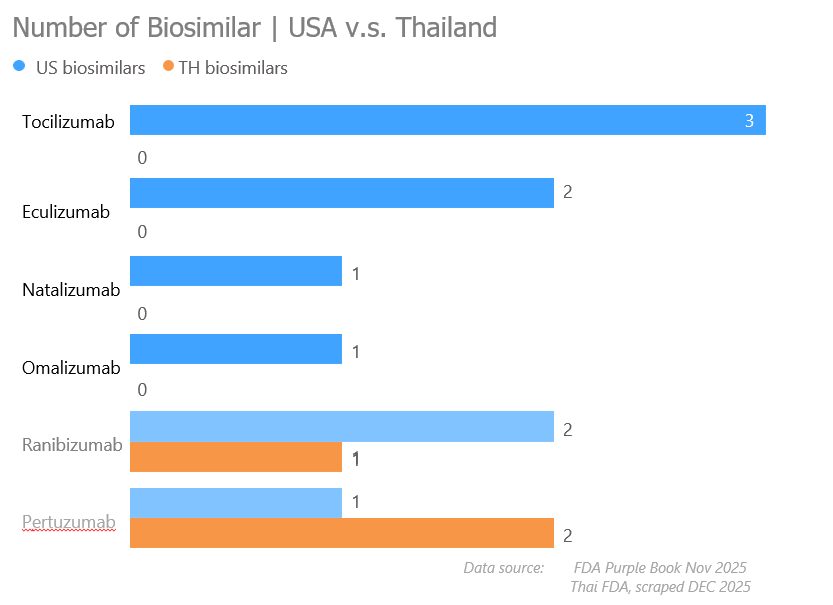
Visualizations helps me create scoring:
| Variable | Scored Value |
| 1. World Top 20 Revenue | 2 pts if yes 0 pts if No |
| 2. High-Impact Disease | 3 pts if High impact 2 pts if Medium impact 1 pt if Specialized impact |
| 3. NLEM Status | 2 pts if NO 0 pts if YES |
| 4. Biosimilar Gap | 3 pts if US Yes; TH No 2 pts if US Yes; 1-2 biosimilar TH 1 pt if US No; TH No or 3 TH 0 pts else |
Formulation for evaluate potential of mAbs:
TMOI = Rev + Tier + NLEM Gap + Biosimilar gap
4. Analyzing
I synthesized all variables into a 10-point scale: the TMOI. Our mAbs for potential market entry and production feasibility are:
- Tocilizumab (9 pts)
- Omalizumab (7 pts)
- Pertuzumab (7 pts)
After identified potential molecules, I would recommend the team to prioritize these three molecules for comprehensive feasibility and production studies.
Claims & Cautions
- Global revenue data is from private industry reports and should be treated as an estimate.
- This impact tiering is a simplified version of NLEM criteria. In a real-world scenario, Cost-Effectiveness and HTA (Health Technology Assessment) would require much deeper modeling.
- Many mAb manufacturers located outside the US are not included in this project.
Project Technical Stack (Hi, recruiters!)
To bring this analysis to life, I utilized a mix of clinical domain knowledge and technical tools:
- Data acquisition: Python (Scrapy) for web scraping Thai FDA data.
- Data engineering: Power Query and Excel for cleaning and joining international datasets.
- Data modeling: Relational database design in Power BI.
- Analytics & Visualization: DAX for scoring logic and Power BI for storytelling.
- Clinical Frameworks: NLEM criteria, and Thai Health Data Center (HDC) data.

Leave a comment